Browse by Organisms Motifs
| M. musculus Motifs of MC12 group | ||||
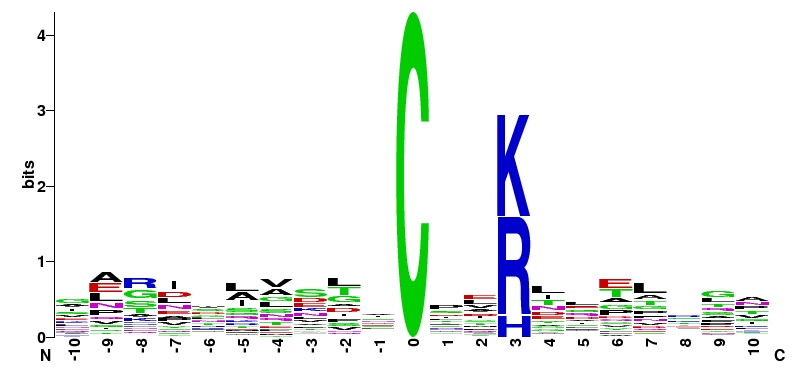 |
| No. | SwissProt ID | Position | S-nitrosylated Peptide | PubMed ID |
| 1 | ACADS_MOUSE | 289 | ALGIAQASLD C AVKYAENRNA | 20925432 |
| 2 | ACADS_MOUSE | 289 | ALGIAQASLD C AVKYAENRNA | 21278135 |
| 3 | ACADS_MOUSE | 289 | ALGIAQASLD C AVKYAENRNA | 22865876 |
| 4 | ACO13_MOUSE | 40 | LVSAAPEKLI C EMKVEEQHTN | 21278135 |
| 5 | ACO13_MOUSE | 74 | VDSISTMALM C TERGAPGVSV | 21278135 |
| 6 | ACO13_MOUSE | 74 | VDSISTMALM C TERGAPGVSV | 22865876 |
| 7 | ACTB_MOUSE | 272 | FQPSFLGMES C GIHETTFNSI | 22178444 |
| 8 | ACTB_MOUSE | 272 | FQPSFLGMES C GIHETTFNSI | 24926564 |
| 9 | ACTB_MOUSE | 272 | FQPSFLGMES C GIHETTFNSI | 18283105 |
| 10 | ACTB_MOUSE | 272 | FQPSFLGMES C GIHETTFNSI | 20837516 |
| 11 | ACTB_MOUSE | 272 | FQPSFLGMES C GIHETTFNSI | 22865876 |
| 12 | ACTN2_MOUSE | 251 | DERAIMTYVS C FYHAFAGAEQ | 22865876 |
| 13 | ADCK3_MOUSE | 265 | ANAERIVSTL C KVRGAALKLG | 21278135 |
| 14 | ADCK3_MOUSE | 638 | AMFEEAYSNY C RMKSGLQ | 21278135 |
| 15 | ADCK3_MOUSE | 638 | AMFEEAYSNY C RMKSGLQ | 22865876 |
| 16 | AIBP_MOUSE | 277 | LNLPSYPDTE C VYRLQ | 21278135 |
| 17 | AMPL_MOUSE | 376 | EGRLILADAL C YAHTFNPKVI | 21278135 |
| 18 | AMPM1_MOUSE | 174 | EEIDHAVHLA C IARNCYPSPL | 21278135 |
| 19 | AP2A1_MOUSE | 941 | IIQTKALQVG C LLRLEPNAQA | 24895380 |
| 20 | AP2A1_MOUSE | 941 | IIQTKALQVG C LLRLEPNAQA | 21278135 |
| 21 | ARBK1_MOUSE | 72 | KLGYLLFRDF C LNHLEEAKPL | 22178444 |
| 22 | ARBK1_MOUSE | 72 | KLGYLLFRDF C LNHLEEAKPL | 17482545 |
| 23 | AT1A1_MOUSE | 374 | VETLGSTSTI C SDKTGTLTQN | 22588120 |
| 24 | AT1A1_MOUSE | 374 | VETLGSTSTI C SDKTGTLTQN | 24895380 |
| 25 | AT1A1_MOUSE | 374 | VETLGSTSTI C SDKTGTLTQN | 21278135 |
| 26 | AT1A1_MOUSE | 374 | VETLGSTSTI C SDKTGTLTQN | 22865876 |
| 27 | AT1A2_MOUSE | 372 | VETLGSTSTI C SDKTGTLTQN | 22588120 |
| 28 | AT1A2_MOUSE | 372 | VETLGSTSTI C SDKTGTLTQN | 21278135 |
| 29 | AT1A2_MOUSE | 372 | VETLGSTSTI C SDKTGTLTQN | 22865876 |
| 30 | AT1A3_MOUSE | 364 | VETLGSTSTI C SDKTGTLTQN | 22588120 |
| 31 | AT1A3_MOUSE | 364 | VETLGSTSTI C SDKTGTLTQN | 24895380 |
| 32 | AT1B3_MOUSE | 191 | LIPDGYPQIS C LPKEENATIA | 22588120 |
| 33 | AT2A2_MOUSE | 349 | VETLGCTSVI C SDKTGTLTTN | 22865876 |
| 34 | ATP4A_MOUSE | 383 | VETLGSTSVI C SDKTGTLTQN | 21278135 |
| 35 | CPSM_MOUSE | 225 | GNPTKVVAVD C GIKNNVIRLL | 20837516 |
| 36 | CPSM_MOUSE | 600 | YALGGLGSGI C PNKETLIDLG | 20837516 |
| 37 | CPT1B_MOUSE | 86 | KVDISMGLVD C IQRCLPERYG | 21278135 |
| 38 | CPT1B_MOUSE | 86 | KVDISMGLVD C IQRCLPERYG | 22865876 |
| 39 | CPT2_MOUSE | 639 | SGRNAREFLH C VQKCLEDMFD | 21278135 |
| 40 | CSDE1_MOUSE | 129 | APGQSPTGSV C YERNGEVFYL | 21278135 |
| 41 | DDX21_MOUSE | 585 | IPEVDLVVQS C PPKDVESYIH | 20925432 |
| 42 | DHB5_MOUSE | 206 | YLNQGKLLDF C RSKDIVLVAY | 22178444 |
| 43 | DHB5_MOUSE | 206 | YLNQGKLLDF C RSKDIVLVAY | 20837516 |
| 44 | DHSA_MOUSE | 89 | GLSEAGFNTA C LTKLFPTRSH | 21278135 |
| 45 | DLDH_MOUSE | 69 | KSAQLGFKTV C IEKNETLGGT | 24895380 |
| 46 | DNPEP_MOUSE | 36 | SPSPFHVVAE C RSRLLQAGFR | 21278135 |
| 47 | DNPEP_MOUSE | 36 | SPSPFHVVAE C RSRLLQAGFR | 22865876 |
| 48 | ECH1_MOUSE | 181 | IGGGVDLVSA C DIRYCTQDAF | 22865876 |
| 49 | ESTD_MOUSE | 206 | ESKWKAYDAT C LVKAYSGSQI | 21278135 |
| 50 | F16P2_MOUSE | 331 | SPEDVQEYLS C VQRNQAGR | 21278135 |
| 51 | F16P2_MOUSE | 331 | SPEDVQEYLS C VQRNQAGR | 22865876 |
| 52 | FAAA_MOUSE | 408 | GDGYRVGFGQ C AGKVLPALSP | 22178444 |
| 53 | FAAA_MOUSE | 408 | GDGYRVGFGQ C AGKVLPALSP | 20837516 |
| 54 | FKBP4_MOUSE | 328 | RLASHLNLAM C HLKLQAFSAA | 21278135 |
| 55 | FLNA_MOUSE | 623 | VEGPSQAKIE C DDKGDGSCDV | 20925432 |
| 56 | FLNA_MOUSE | 631 | IECDDKGDGS C DVRYWPQEAG | 20925432 |
| 57 | FLNB_MOUSE | 1280 | ITNPSGASTE C FVKDNADGTY | 20925432 |
| 58 | HNRPL_MOUSE | 258 | LNGADIYSGC C TLKIEYAKPT | 22588120 |
| 59 | HNRPL_MOUSE | 258 | LNGADIYSGC C TLKIEYAKPT | 20925432 |
| 60 | HXK2_MOUSE | 909 | KGAALITAVA C RIREAGQR | 21278135 |
| 61 | HXK2_MOUSE | 909 | KGAALITAVA C RIREAGQR | 22865876 |
| 62 | ILK_MOUSE | 346 | SMADVKFSFQ C PGRMYAPAWV | 21278135 |
| 63 | ISC2A_MOUSE | 136 | GLQVHVVVDA C SSRSQVDRLV | 21278135 |
| 64 | ISCA2_MOUSE | 56 | GEGQIRLTDS C VQRLLEITEG | 21278135 |
| 65 | LEG1_MOUSE | 61 | NAHGDANTIV C NTKEDGTWGT | 20925432 |
| 66 | LEG1_MOUSE | 61 | NAHGDANTIV C NTKEDGTWGT | 21278135 |
| 67 | LEG1_MOUSE | 61 | NAHGDANTIV C NTKEDGTWGT | 22865876 |
| 68 | LHPP_MOUSE | 226 | IVGDVGGAQQ C GMRALQVRTG | 21278135 |
| 69 | MK01_MOUSE | 159 | KPSNLLLNTT C DLKICDFGLA | 22178444 |
| 70 | MK01_MOUSE | 159 | KPSNLLLNTT C DLKICDFGLA | 19101475 |
| 71 | MK01_MOUSE | 159 | KPSNLLLNTT C DLKICDFGLA | 19483679 |
| 72 | MPCP_MOUSE | 232 | QIPYTMMKFA C FERTVEALYK | 21278135 |
| 73 | MYOM1_MOUSE | 1601 | NEKPLTSDDH C SLKFEAGKTA | 21278135 |
| 74 | MYOM1_MOUSE | 1601 | NEKPLTSDDH C SLKFEAGKTA | 22865876 |
| 75 | NAMPT_MOUSE | 397 | LQKLTRDLLN C SFKCSYVVTN | 21278135 |
| 76 | NDUS1_MOUSE | 554 | MLFLLGADGG C ITRQDLPKDC | 21278135 |
| 77 | NDUS1_MOUSE | 554 | MLFLLGADGG C ITRQDLPKDC | 22865876 |
| 78 | NLRX1_MOUSE | 644 | HNRAVLAQLG C PIKNLDALEN | 21278135 |
| 79 | NMI_MOUSE | 183 | QTRDKLELSF C KSRNGGGEVE | 20925432 |
| 80 | NNRE_MOUSE | 277 | LNLPSYPDTE C VYRLQ | 22865876 |
| 81 | NNTM_MOUSE | 936 | SIVITPGYGL C AAKAQYPIAD | 22865876 |
| 82 | ODBB_MOUSE | 186 | GSLTIRAPWG C VGHGALYHSQ | 22865876 |
| 83 | P5CR3_MOUSE | 49 | LASAPTDNNL C HFRALGCQTT | 21278135 |
| 84 | PCBP1_MOUSE | 54 | GARINISEGN C PERIITLTGP | 20925432 |
| 85 | PCBP1_MOUSE | 54 | GARINISEGN C PERIITLTGP | 21278135 |
| 86 | PCBP2_MOUSE | 54 | GARINISEGN C PERIITLAGP | 20925432 |
| 87 | PCBP2_MOUSE | 54 | GARINISEGN C PERIITLAGP | 21278135 |
| 88 | PCCB_MOUSE | 271 | RAFDNDVDAL C NLREFFNFLP | 21278135 |
| 89 | PCCB_MOUSE | 271 | RAFDNDVDAL C NLREFFNFLP | 22865876 |
| 90 | PCNA_MOUSE | 135 | QLGIPEQEYS C VIKMPSGEFA | 19483679 |
| 91 | PP2AA_MOUSE | 251 | HQLVMEGYNW C HDRNVVTIFS | 21278135 |
| 92 | PPIA_MOUSE | 115 | PNTNGSQFFI C TAKTEWLDGK | 21278135 |
| 93 | PPIA_MOUSE | 115 | PNTNGSQFFI C TAKTEWLDGK | 22865876 |
| 94 | PPIF_MOUSE | 156 | PNTNGSQFFI C TIKTDWLDGK | 21278135 |
| 95 | PPIF_MOUSE | 156 | PNTNGSQFFI C TIKTDWLDGK | 22865876 |
| 96 | PREB_MOUSE | 161 | FSNEPLQKVV C FNHDNTLLAT | 20925432 |
| 97 | PSD11_MOUSE | 202 | SARTTANAIY C PPKLQATLDM | 21278135 |
| 98 | PSD11_MOUSE | 202 | SARTTANAIY C PPKLQATLDM | 22865876 |
| 99 | PUR9_MOUSE | 434 | AVKYTQSNSV C YAKDGQVIGI | 21278135 |
| 100 | SBP1_MOUSE | 31 | GPREEIVYLP C IYRNTGTEAP | 22178444 |
| 101 | SBP1_MOUSE | 31 | GPREEIVYLP C IYRNTGTEAP | 21278135 |
| 102 | SBP1_MOUSE | 31 | GPREEIVYLP C IYRNTGTEAP | 20837516 |
| 103 | SBP1_MOUSE | 31 | GPREEIVYLP C IYRNTGTEAP | 22865876 |
| 104 | SDHA_MOUSE | 89 | GLSEAGFNTA C LTKLFPTRSH | 22588120 |
| 105 | SDHA_MOUSE | 89 | GLSEAGFNTA C LTKLFPTRSH | 22865876 |
| 106 | SKP1_MOUSE | 160 | EAQVRKENQW C EEK | 22178444 |
| 107 | SKP1_MOUSE | 160 | EAQVRKENQW C EEK | 19101475 |
| 108 | SKP1_MOUSE | 160 | EAQVRKENQW C EEK | 21278135 |
| 109 | SODC_MOUSE | 7 | ----MAMKAV C VLKGDGPVQG | 24895380 |
| 110 | SOX_MOUSE | 333 | NTPDEHFILD C HPKYDNIVIG | 22178444 |
| 111 | SOX_MOUSE | 333 | NTPDEHFILD C HPKYDNIVIG | 20837516 |
| 112 | SYG_MOUSE | 451 | TSYGWIEIVG C ADRSCYDLSC | 22865876 |
| 113 | SYIM_MOUSE | 883 | PGLEEAVESA C AMRDSFLGSI | 21278135 |
| 114 | SYIM_MOUSE | 883 | PGLEEAVESA C AMRDSFLGSI | 22865876 |
| 115 | SYVC_MOUSE | 1184 | GCAVAVASDR C SIHLQLQGLV | 20925432 |
| 116 | TITIN_MOUSE | 2879 | EYTAMVGQLE C KAKLFVETLH | 22865876 |
| 117 | TITIN_MOUSE | 33667 | TATNTAGSTS C QAHLQVERLR | 21278135 |
| 118 | TNR21_MOUSE | 211 | KPGTKETDNV C GMRLFFSSTN | 22865876 |
| 119 | TXLNB_MOUSE | 652 | SEGDSAVVPG C ESREQPPPEV | 21278135 |
| 120 | UB2V1_MOUSE | 71 | ENRIYSLKIE C GPKYPEAPPS | 21278135 |
| 121 | UBA1_MOUSE | 340 | HIGFQALHQF C ALHNQPPRPR | 20837516 |
| 122 | XPO2_MOUSE | 939 | AQSLHKLSTA C PGRVPSMVST | 20925432 |
